|
# # # # Connecting genetics and biology is complicated. Researchers around the world have struggled to determine what each functional region of DNA is doing individually, let alone in combination with other regions. And sometimes when the output of combinations is examined, the results can be unexpected. Recently, researchers looked at the consequences of having a particular combination of Parkinson’s-associated risk factors… and they were rather surprised by the results In today’s post, we will review the report presenting their results and consider the potential implications of the findings. # # # # |

A while back, I became a little obsessed with peacock feathers.
I didn’t start collecting them and wearing them on Saturday night or anything like that. Rather, I just got really fascinated with how they develop. Each individual feather, that is.
I mean, look at them:

Like all organisms, they are wondrous feats of nature and biology – particularly the jewel-toned ocelli (plural) or eyespots (the vivid circular patterns that seem evenly spread along each feather).
Each ocellus (singular) is created via a combination of individual strands of the larger feather. And each strand is further made up of tiny individually coloured segments. When you get really up close and personal with those eyespots, they look like this:

My obsession centered around “the how”.
How does each strand of the feather know when to start some blue or gold colouration (and when to stop) along those strands? And how do the individual strands coordinate and match up so perfectly to create the marvelous image of the ocellus?
This type of question applies to many areas of biology (for example, how does a regenerating tail of a gecko know when to stop growing?), but remember that at the end of each mating season, the peacock sheds (or molts) its feathers. So these carefully coordinated feathers have to re-grow each year!
Tell me that that is not remarkable.
Remarkable, but what does this have to do with Parkinson’s?
Well, my assumption was that the development of each feather and the ocelli would involve a very complicated combination of genetics. But given our limited current understanding of genetics, I doubted that we would have the foggiest idea of how peacock feathers develop.
I thought we know a lot about genetics. What do you mean “our limited current understanding of genetics”?
With the development of DNA sequencing technology, we have learned a great deal about the genetics of individual organisms over the last 20 years. It has been a golden age for biomedical research – one that is slowly transforming the world we live in.
But, while all of these advances in our knowledge have been encouraging, when we actually discuss “genetics”, most researchers will tell you that we are still only scratching the surface in terms of understanding the true complexity of DNA in an creature like humans.
Particularly in terms of how combinations of specific genetic interactions give rise to a particular trait (or observable characteristic).
What do you mean “combinations of specific genetic interactions”?
In almost every cell of your body you have DNA (or Deoxyribonucleic acid). It is a molecule that is composed of two chains which coil around each other, forming the famous “double helix” shape.

The information stored in DNA provides the template or the instructions for making (and maintaining) a particular organism. All of the necessary details are encoded in that amazing molecule.
All along the long chains of DNA (there is over 2 meters of DNA in each cell in humans), there are regions that can be ‘read’ and transcribed into RNA (RNA being the template of DNA that is used for the production of protein).
Each of these regions is called a gene.
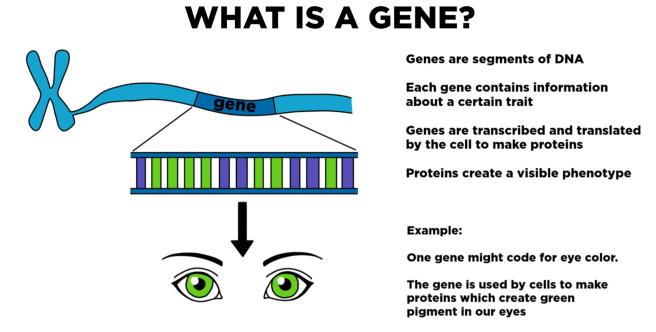
In humans, there are estimated to be at least 46,831 genes (plus another 2300 micro-RNA genes) (Source).
That’s a lot.
And for the vast majority of those genes we have very limited information. Researchers have previously tended to adopt herd mentality and focused their studies on the genes that are most in vogue for a particular area of biology (think, SNCA (alpha synuclein) for Parkinson’s). This has resulted in us knowing a lot about very few genes.
So we know a lot of about a few key genes in terms of particular traits or medical conditions, but little about the influence that many other genes might be having. And most of our current understanding of the key genes has come from investigating the function of those individual genes. It is very rare that they are studied in combination with other genes.
Why is that?
If you want to know the function of a gene, the basic approach in experimental biology is to take a model system (such as a cell in culture or a fly, or a mouse) and you do one of two things:
- You remove the normal copy of a gene – and therefore observe what happens in its absence (this can be referred to as a Loss of Function (LOF) study)
- You enhance the activity of the gene – increase the activity and record what happens (often referred to as a Loss of Function (LOF) study)
Basically, reduce or boost the activity of a gene. Sound simple right?
Unfortunately it is not. Firstly, context is key. Different results can be seen in different tissues and model systems (funnily, flies are rather different to humans). Plus the timing of the experiment is important – many genes have different roles in cellular behaviour at different time points in the life cycle of a cell (for those interested in this exploring this topic further, click here for a good review).
So we have learned a lot about a few genes in many different model systems and at different stages of the cell cycle. But the great “dark matter” aspect of this type of research is what influence all of the other unaffected genes might be having in these experiments.
Some genes may compensate for the loss of a particular gene, while tiny unknown variations in dozen of genes in a cell might impact the results of a particular study (giving potentially contradicting results). The effect that combinations of genetic interactions could be having is next level in terms of complexity.
A better understanding of how combinations of genes affect certain traits would greatly enhance our understanding of biology, particularly in terms of medical conditions like Parkinson’s.
|
# RECAP #1: Our current understanding of genetics and biology is largely based on studies exploring the activity of individual genes. But in reality, it is probably combinations of genes that impact particular traits (or characteristics). # |
Ok, so genetics is complicated. How can we learn more about the genetics of Parkinson’s then?
Well, setting up large longitudinal cohort studies and investigating the relationship between particular combinations of genetic risk factors for PD and their clinical characteristics is one way.
And recently there was a new report published that demonstrated a rather surprising result.
What was it?
This is the report in question:

Authors: Ortega RA, Wang C, Raymond D, Bryant N, Scherzer CR, Thaler A, Alcalay RN, West AB, Mirelman A, Kuras Y, Marder KS, Giladi N, Ozelius LJ, Bressman SB, Saunders-Pullman R.
Journal: JAMA Netw Open. 2021 Apr 1;4(4):e215845.
PMID: 33881531 (This report is OPEN ACCESS if you would like to read it)
In this study, the researchers collected data from multiple longitudinal studies that were following people with Parkinson’s over time. These studies included:
- the Mount Sinai Beth Israel Biomarker study,
- the Parkinson Disease Biomarker Program,
- the Harvard Biomarkers Study,
- Ashkenazi Jewish-LRRK2-Consortium,
- Parkinson Progression Marker Initiative,
- SPOT-PD studies,
In total, they had genetic and clinical data from 1193 people with Parkinson’s. Of these, there were 128 individuals who carried a GBA gene variant that is associated with PD. There were also 155 individuals who had variants in their LRRK2 gene that are associated with PD. But of particular interest to our discussion here today were the 21 individuals in the study who had a combination of genetic variations in both their LRRK2 and GBA genes. In other words, these 21 people carried two genetic risk factors for Parkinson’s.
The remaining 889 cases had what is known as idiopathic Parkinson’s (this is spontaneous form of the condition that cannot be attributed to a specific genetic risk factor).
Quick question: What is the GBA gene?
We have written a lot about the GBA gene on this website – click here to read more about GBA and GBA-associated Parkinson’s.
Ok. Another question: What is the LRRK2 gene?
We have written a lot about the LRRK2 gene on this website – click here to read more about LRRK2 and LRRK2-associated Parkinson’s.
Before we continue, however, understand that a number of cohort studies have been conducted on people with GBA-associated Parkinson’s and people with LRRK2-associated Parkinson’s, and there are two basic patterns:
- People with GBA-associated Parkinson’s typically have a more rapid course of disease progression (although there is a lot of variability between cases)
- People with LRRK2-associated Parkinson’s typically have a slower course of disease progression (although there is a lot of variability between cases)
And when the researchers (who conducted the study we are reviewing today) looked at the clinical scores of the GBA-associated Parkinson’s and LRRK2-associated Parkinson’s cases in their data set, this is exactly what they saw: over time, cases of GBA-associated Parkinson’s were more rapid than idiopathic PD, and LRRK2-associated Parkinson’s cases were slower than idiopathic PD.
What about the people with both LRRK2 and GBA genetic variants?
So this is where the study gets really interesting.
You see, basic reasoning would suggest that if you have two genetic risk factors for a particular medical condition, you would probably have a case of that disease that is twice as bad as normal. Right?
But that is not what the researchers found in this study.
Rather, they reported that the clinical measures over time suggested that people with both LRRK2 and GBA variants had a very similar rate of progression to the LRRK2-associated Parkinson’s in terms of their motor symptom progression. Their rate of progression was slower than idiopathic PD.
In the image below, the upward trajectory of the lines illustrates a worsening of motor symptoms, and you can see that the yellow line of the GBA-associated PD cases is steeper than the black line of the idiopathic PD cases, suggesting faster progression. And the light blue line of the LRRK2-associated PD cases is less steep than the idiopathic PD cases, indicating slower progression. The grey line of the LRRK2 and GBA variant combination group exhibits a very similar gradient to the the light blue line of the LRRK2-associated PD cases:

And the researchers also saw this pattern in measures of cognitive capacity (as measured by the MoCA cognitive assessment test), see the image below (where a downward trajectory indicated a worsening of capacity):

The researchers concluded that the combination of LRRK2 and GBA variants did not have a combined deleterious effect on the progression of Parkinson’s symptoms. Rather, LRRK2 variant may be biologically masking the effects of GBA variations.
|
# RECAP #2: A new study reported that people with genetic mutations in both their LRRK2 and GBA genes does not exaggerate the progression of Parkinson’s symptoms. Rather the course of their Parkinson’s is more in line with people who have Parkinson’s-associated LRRK2 variants alone, than GBA variants. # |
Interesting. Is this the first time this has been reported?
Actually, no.
Two studies that I am aware of have presented some data suggesting a “protective effect” (or possibly a “dominant effect”) in PD carriers of both GBA + LRRK2 mutations (Click here and here to read more about these studies).
Any idea as how this effect could be explained?
LRRK2 variants have been reported to increase levels of GCase activity in blood (GCase is the enzyme that is produced from the RNA of the GBA gene), which could partly explain the less severe course of the combination of LRRK2 + GBA variants.
For example, there was this paper in 2015:

Authors: Alcalay RN, Levy OA, Waters CC, Fahn S, Ford B, Kuo SH, Mazzoni P, Pauciulo MW, Nichols WC, Gan-Or Z, Rouleau GA, Chung WK, Wolf P, Oliva P, Keutzer J, Marder K, Zhang X.
Journal: Brain. 2015 Sep;138(Pt 9):2648-58.
PMID: 26117366 (This report is OPEN ACCESS if you would like to read it)
In this study, the researchers found that individuals carrying the LRRK2 G2019S variant (one of the most common LRRK2 variants associated with Parkinson’s) had higher enzymatic activity than non-carriers (with and without PD), and this was associated with longer disease duration.
Other Parkinson’s-associated LRRK2 variants (such as p.M1646T) also have higher levels of GCase activity in blood cells (Click here to read more about this). Whether these results are specific to blood and not brain still needs to be determined (GCase activity has been reported to be reduced in dopamine neurons derived from PD patients with LRRK2 mutations – Click here to read more about this), but it could indicate a possible mechanism for the effect being observed in individuals with LRRK2 + GBA variants.
How might these results effect Parkinson’s research?
Well, in clinical trials focused on disease modification in GBA-associated Parkinson’s, the investigators will need to screen for individuals who have both GBA and LRRK2 genetic variants. If the combination of GBA and LRRK2 variants results in a slower progression, this means that these individuals (rare as they may be) could potentially impact the outcome of the study, and would need to be noted or possibly excluded.
For example, efforts are currently being made to set up a large trial of the respiratory medication ambroxol, which is being repurposed for Parkinson’s by Cure Parkinson’s (Click here to read a previous SoPD post about this topic).

Participants who will take part in the future study will be screened for GBA genetic variants (as the researchers are keen to test this treatment in a GBA-associated PD cohort), but they may also need to be screened for LRRK2 variants so that the additional genetic mutation does not potentially influence the result.
How can I find out if I have a GBA or LRRK2 genetic variant?
There are now numerous genetic testing services available that look for GBA and LRRK2 variants.
Here in the UK, there is the PD Frontline initiative which is screening for GBA and LRRK2 genetic mutations.

|
# An important note regarding genetic testing: Learning about one’s genetic status is a personal choice, but it is not something that should be taken lightly and rushed into. Discovery of unexpected/unwanted information (such as APOE4 or BRCA1 status, parentage, etc) can be dramatic – both for the individual and their family. The whole genetic testing process needs to be handled carefully, and should always be done with genetic counseling. # |
So what does it all mean?
There is a lot of variability between cases of Parkinson’s, which could be partly explained by some of the variations in our DNA that make us each unique. Combinations of tiny errors that give us different skin and eye colours, different height and shape, different vulnerabilities to particular aliments. These are the things that make us all individuals.
But new research into the combination of two different types of Parkinson’s risk factors has provided some intriguing insights into the biology underlying the condition, raising new questions and giving rise to new research and issues to be aware of.
It is important to appreciate that not all LRRK2 genetic variants associated with Parkinson’s are the same – 7 of the reported LRRK2 mutations have been identified as pathogenic (R1441G, R1441C, R1441H, Y1699C, G2019S, R1628P, G2385R and I2020T) which are all located in different functional regions of the LRRK2 gene (Source). Nor are all GBA mutations the same (Source). So this new research must be taken with a pinch of salt – as we have just said, there is a lot of variability between cases. But the findings of the study do point towards the need for greater awareness and understanding of the potential impact of genetic status could have on outcomes, which could (in theory) have significant effect on the outcomes of clinical trials (Source).
For those interested in learning more about LRRK2- and GBA-associated Parkinson’s and efforts to target them in a more “precision medicine” approach, click here to read a good recent review on the topic.
And now back to the peacocks…
Reflections on the feather of peacocks
“I remember well the time when the thought of the eye made me cold all over, but I have got over this stage of the complaint, and now small trifling particulars of structure often make me very uncomfortable. The sight of a feather in a Peacock’s tail, whenever I gaze at it, makes me sick”
– Charles Darwin (Source)
The male Indian peafowl (Pavo cristatus) starts shaking its tail feathers towards the end of the summer, and the stunning plumage gradually falls off. This process is called molting, and it is driven by hormones which increase at the end of the mating season.

SIDE NOTE: Curiously, if you castrate a peacock it will have a limited impact on the elaborate plumage but ovariectomizing a peahen will cause her to develop a male-type plumage (Source).
Over the next 7 months, the peacock (the male peafowl, not to be confused with the more demure female peahen) will grow a new train of feathers. The train can extend more than 5 feet (1.7 meters) in length, and reaches its peak development when the peacock is around age 6 years of age (15-20 years is average life span).

During the mating season, peacocks will cluster together in groups called leks. Peahens (the females) will then go “window-shopping” by strolling through the lek. To get noticed, the peacocks will raise and spread that trains. They will also shake their plumes (ornithologists call this shivering).
Peacock tail feathers develop to ensure that they reveal each ocellus whenever the bird fans them out – there is very little overlapping. The feather has a central shaft, which is lined by rows of smaller barbs that contain minute barbules (projecting filaments).

These barbules are made up smaller segments of melanin interlaced with keratin.

This video provides further context on the structure of peacock feathers:
To the observer, the ocelli appear in different shades of blues and greens, but tail feathers are actually pigmented brown and the iridescent colours of the feathers are created by structural coloration.
First observed by Robert Hooke and Isaac Newton, structural coloration is production of colour by microscopically structured surfaces fine enough to interfere with visible light, sometimes in combination with pigments (Source).

In the case of the peacock feather, the different angles of the segments of the barbules catch and reflect the sunlight to create the iridescence observed by the eye.
The genetics of the peacock feather is less well understood than human genetics. A recent genetic analysis has provided some insights into the development of the feathers:

Authors: Jaiswal SK, Gupta A, Saxena R, Prasoodanan VPK, Sharma AK, Mittal P, Roy A, Shafer ABA, Vijay N, Sharma VK.
Journal: Front Genet. 2018 Sep 19;9:392.
PMID: 30283495 (This report is OPEN ACCESS if you would like to read it)
In this study, the researchers conducted a comparative genetic analysis of the peacock with five related bird types. When they analysed the approximately 2,100 genes involved in feather development, the investigators found that the peacock genome differed from the other species in terms of activators of feather development (such as FGF, Wnt/β-catenin and TGF-β) and inhibitors (including BMP and Notch-delta) show sequence divergence in peacocks.
The study provides some insights into the development of peacock feathers, but – like Parkinson’s – a better understanding will only come from further analyses of combinations of genes and how their interactions impact the feather.
And then there are albino peacocks:

You also have “India Blue Pied” peacocks, which have 30-50 percent white feathers replacing the coloured feathers:

Like I said, I kinda got a little obsessed with the peacock feather.
Back to more Parkinson’s research in the next post.

Creative Commons Attribution 4.0 International License
You can do whatever you like with it!
EDITOR’S NOTE: The information provided by the SoPD website is for information and educational purposes only. Under no circumstances should it ever be considered medical or actionable advice. It is provided by research scientists, not medical practitioners. Any actions taken – based on what has been read on the website – are the sole responsibility of the reader. Any actions being contemplated by readers should firstly be discussed with a qualified healthcare professional who is aware of your medical history. While some of the information discussed in this post may cause concern, please speak with your medical physician before attempting any change in an existing treatment regime.
Further, the author of this post is an employee of Cure Parkinson’s. The Trust is a supporter of both the ambroxol clinical trial and the PD Frontline project that were discussed in this post. Cure Parkinson’s has not asked for this post to be written. The post has been written by the author solely for the purpose of sharing what the author considers interesting information.
The banner for today’s post was sourced from Marketwatch


